Basic concepts
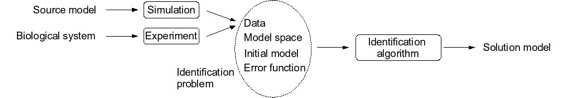
The figure gives an overview of the main concepts involved. The task we are considering is to create an ODE model from time series data with an identification algorithm. The time series data can either come from experiments on a real system or be simulated from a known source model. Before we can enter our problem in an identification algorithm we need to supply additional information to define an identification problem, the details of which is discussed in the next section. If the identification problem is well formulated and the identification algorithm successful, the solution model should be similar to the source model.
Defining identification problems as optimization problems
Unfortunately, many problems considered in the literature are not easy to reconstruct. For example, important information may be missing and/or be scattered in different places. As a consequence, no standard problems are available for testing, making development of algorithms difficult. Comparisons of different algorithms are almost impossible to do. An underlying reason for this is that there has been no established way to describe identification problems when both parameters and structure are unknown, and no standard way to represent such problems in files.
The first problem can be solved by defining the identification unambiguously as an optimization problem. This has several important advantages. First, the identification problem then has a solution (or possibly several solutions) that is independent of any algorithm for solving such problems. Secondly, the task of modelling an identification problem, which is an interesting problem in its own right, becomes independent of the task of solving the problem.
To define an identification problem with unknown structure and parameters, we require the following information:
- Time series data from one or several experiments.
- A model space describing the allowed forms of the right hand side of the ODEs.
- An initial model specifying a known prior information about parts of the structure.
- An error function specifying how well any given model fits with the data. This function must include some notion of model complexity.
At present, the types of ODEs we are considering are chemical rate reactions and S-systems. Previous research on identification algorithms has mostly been concerned with S-systems, but we have also designed a number of new benchmark problems using chemical rate reactions which are more common for biochemical modelling.
For more details about how to define the identification problems, see the original benchmark publication and the documentation.
Introduction to the identification problem file format
We have developed a file format used for all benchmark problems, that can hold the information outlined above. An identification problem can then be represented in a single file. Note that we describe optimization problems and not models, so formats such as SBML cannot be used for this purpose. A short extract of the file format is shown below:
// VARIABLES variable_1 has name = x1 is inputVariable variable_2 has name = x2 is inputVariable variable_3 has name = x3 is dependent ... // MODEL SPACE OF VARIABLE 3 possibleReaction_3 of variable_3 has type = biMolecularMassAction has spaceOfVariable X1 = memberOfSet_2 has spaceOfVariable X2 = memberOfSet_1 has rangeOfParameter k1 = range_1 ... // EXPERIMENT 1 sample_1 of experiment_1 has time = 0.00 has variable_ = 3.00 2.00 ... has sdev of variable_ = 0.00 0.00 ... ...
In the example, some variables are first defined. In the next part a possible term of the ODE for variable 3 is given. Finally, some data is given. Complete files for all benchmark problems are available on this site. Please note that the example does not at all show all features of the format or how we represent different problems.
For more details about the format, see the documentation. Note that both the definition of identification problems outlined above, as well as the file format, will be extended over time to enable representation of more complex identification problems.